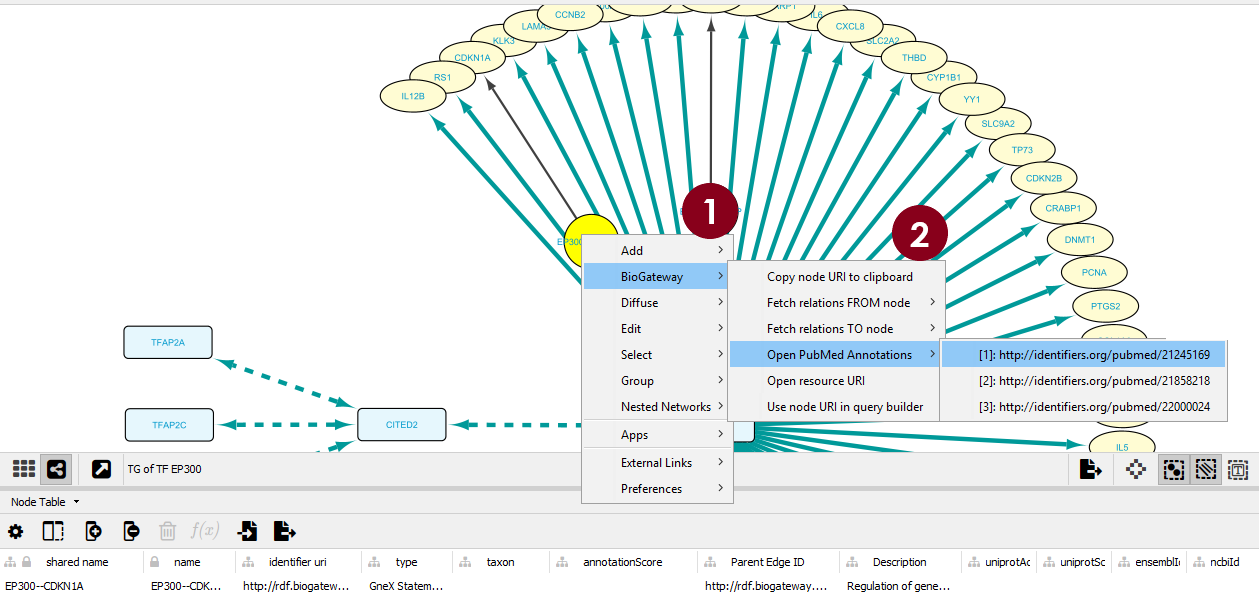
Use Case 2 HOW TO FIND DNA-TF / COACTIVATOR-TF ASSOCIATIONS THAT REGULATE TG
Biological question
Which DNA binding Transcription Factors (dbTFs) interact with a particular coactivator and regulate the transcription of target genes (TGs)?
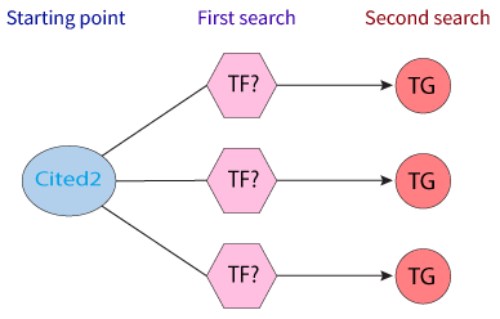
The transcriptional coactivator CITED2 (Cbp/p300-interacting transactivator 2) is involved in many biological processes, such as cell differentiation and development of several tissues, (e.g., cardiac, adrenal, and neural tissues) (PMID: 21764344). Regulation of gene transcription can be accomplished through the interaction of a DNA binding TF (dbTF) with coactivator proteins (coTF), with the dbTF interacting with these coactivators important for the correct functioning of the target genes.
QUERY AND RESULTS
The specific questions are described in a stepwise manner in the lines of the Query Builder and the BioGateway options allow a specific further query.
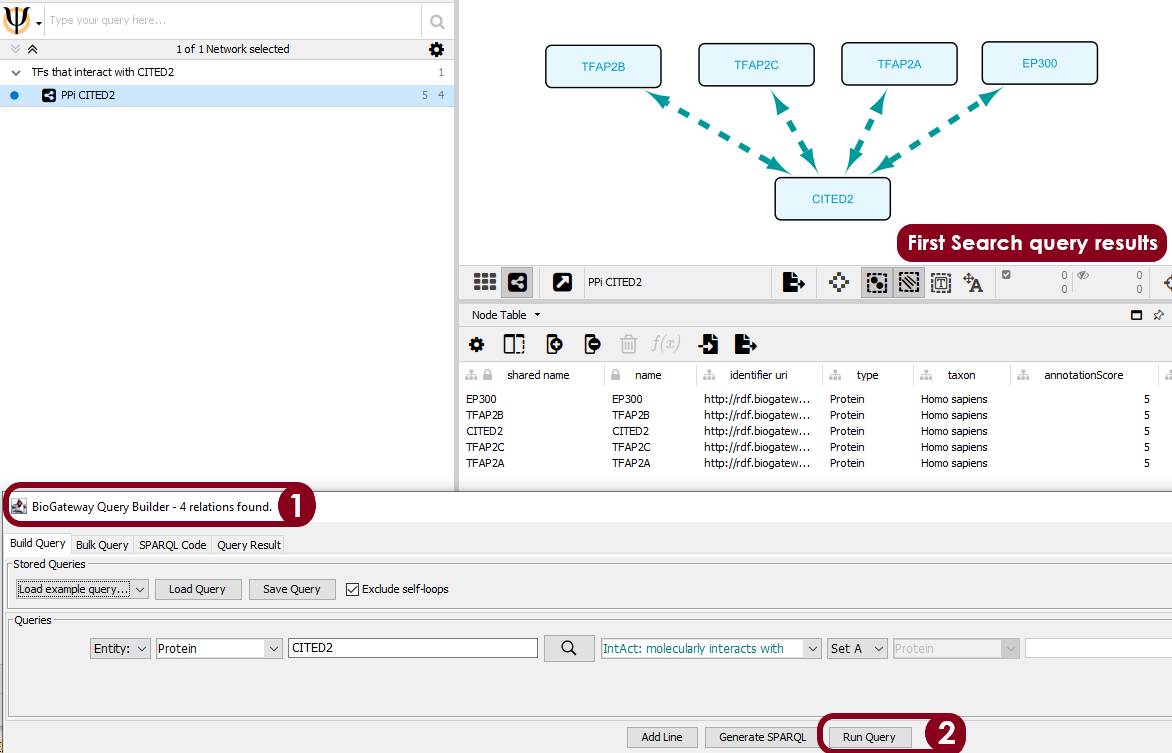
Figure 1. BioGateway Query Builder and First search results
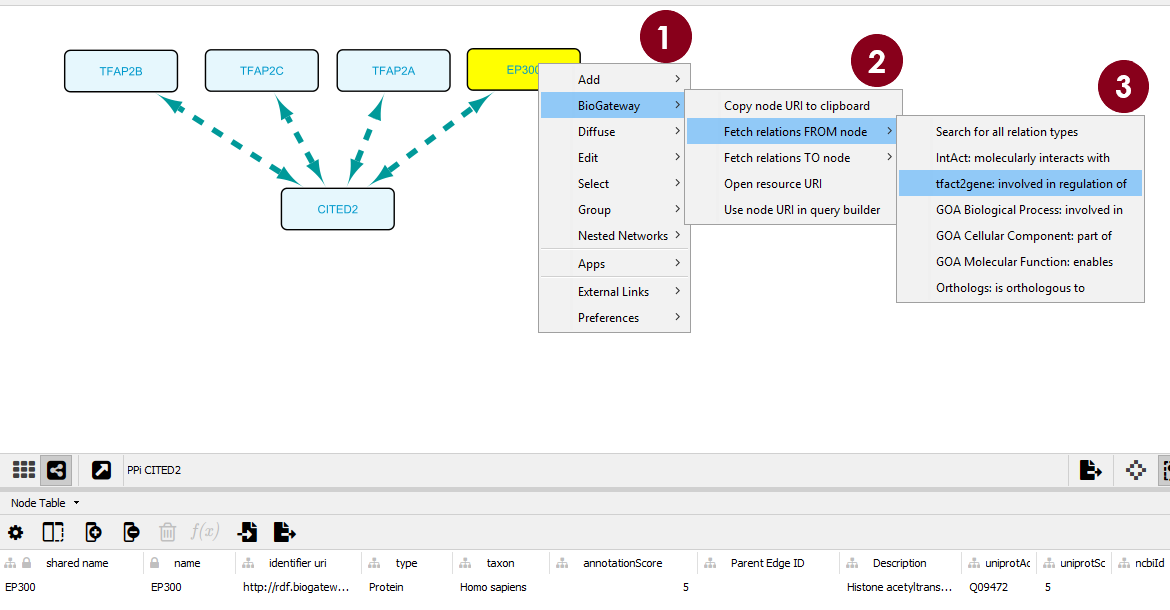
Figure 2. Steps to find the Second search results

Figure 3. Visualization of the Second search results